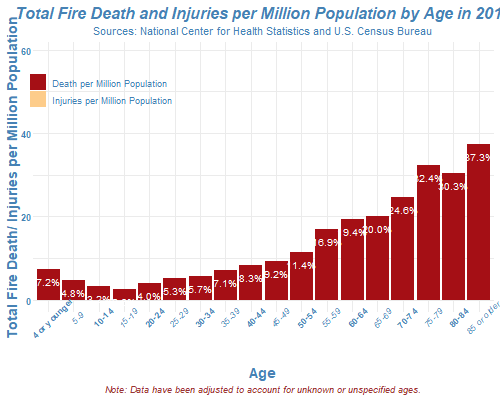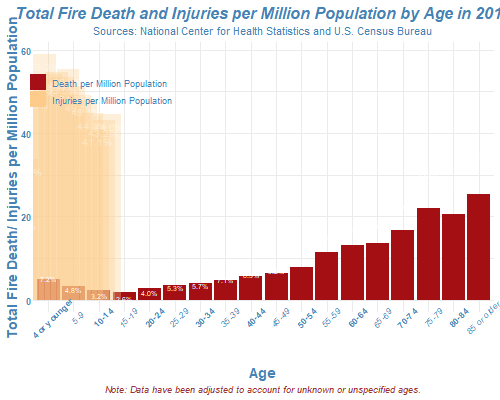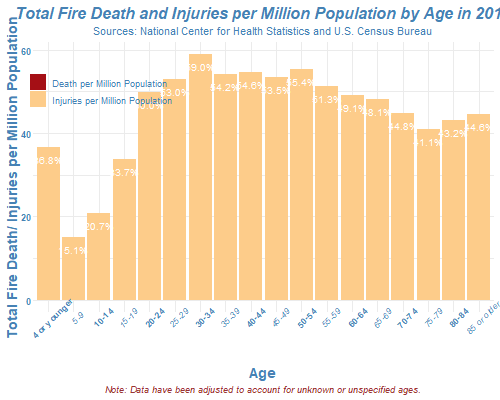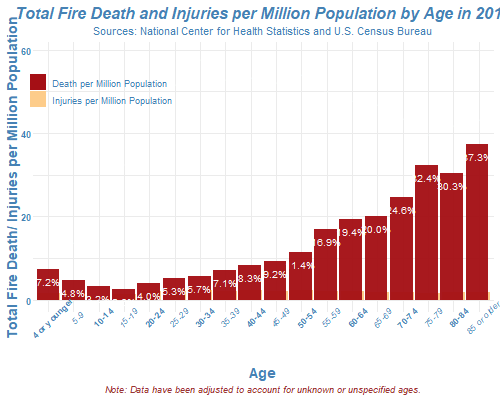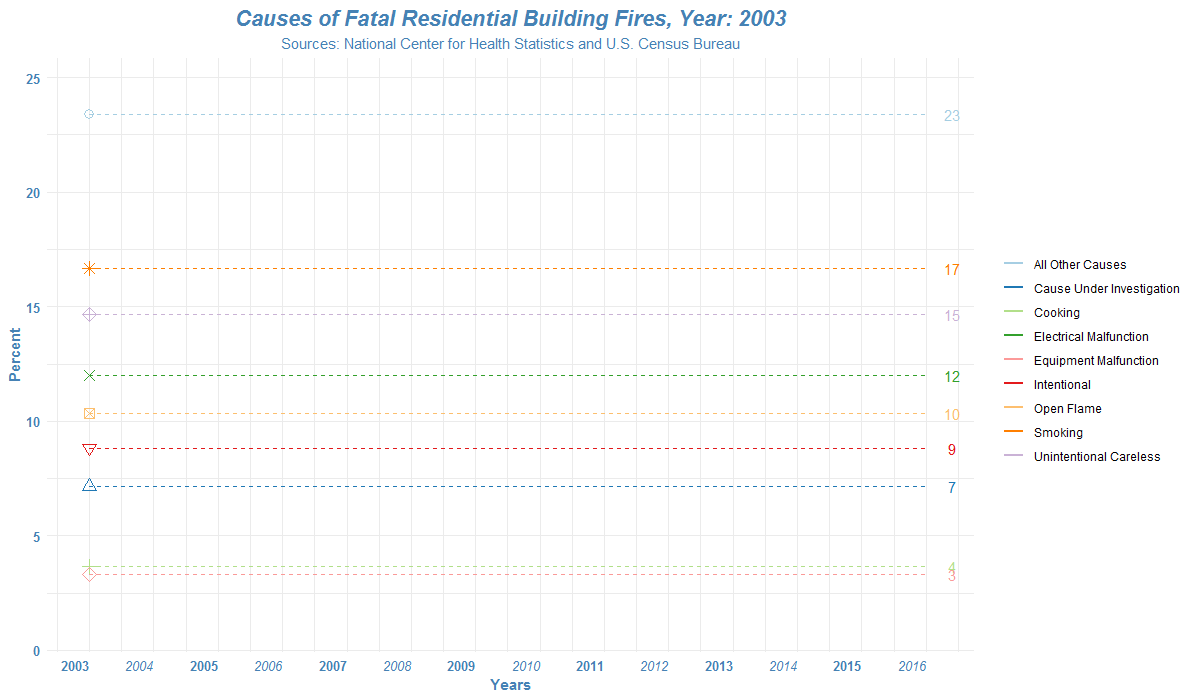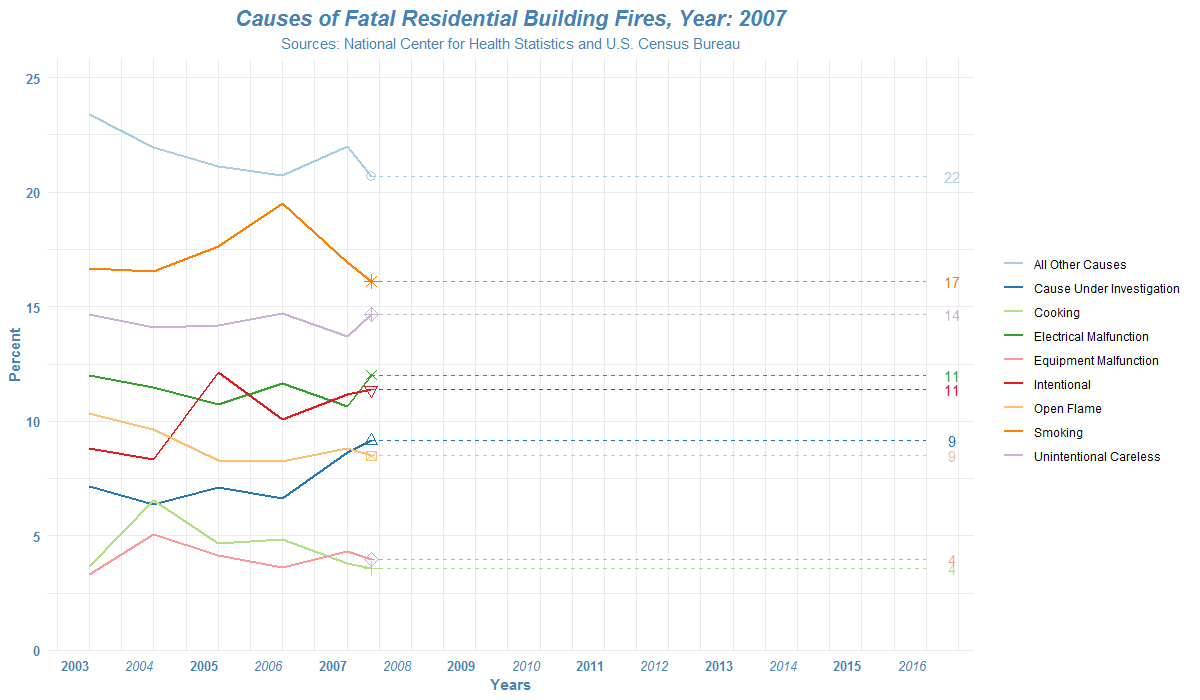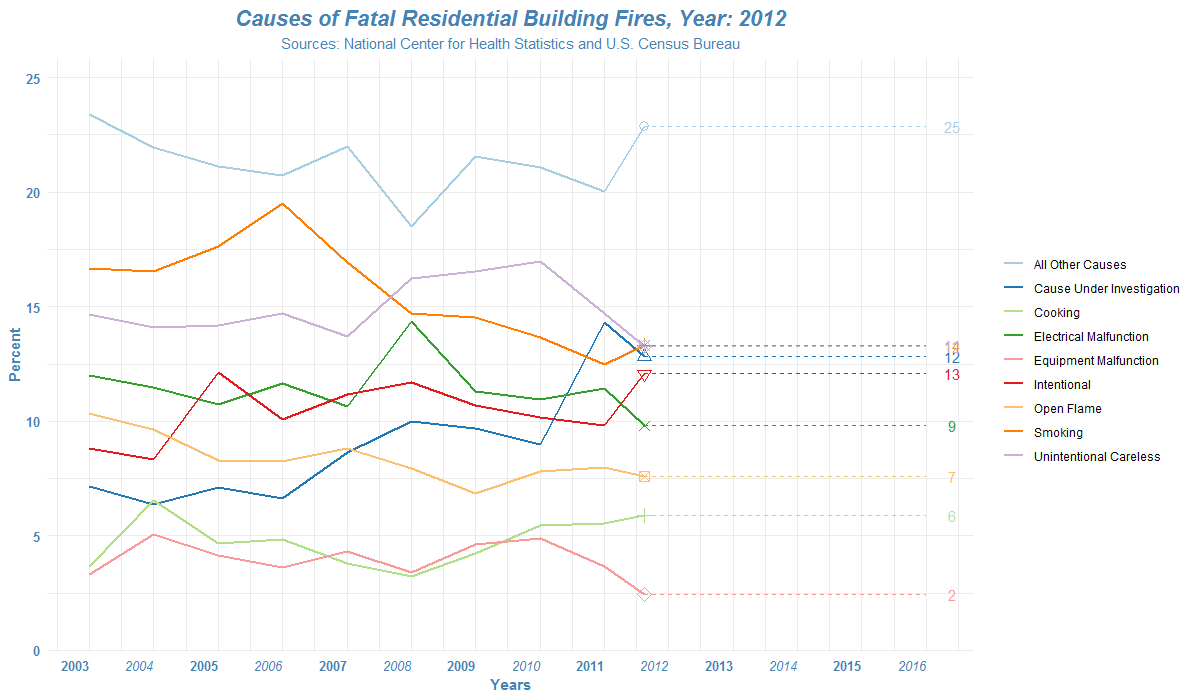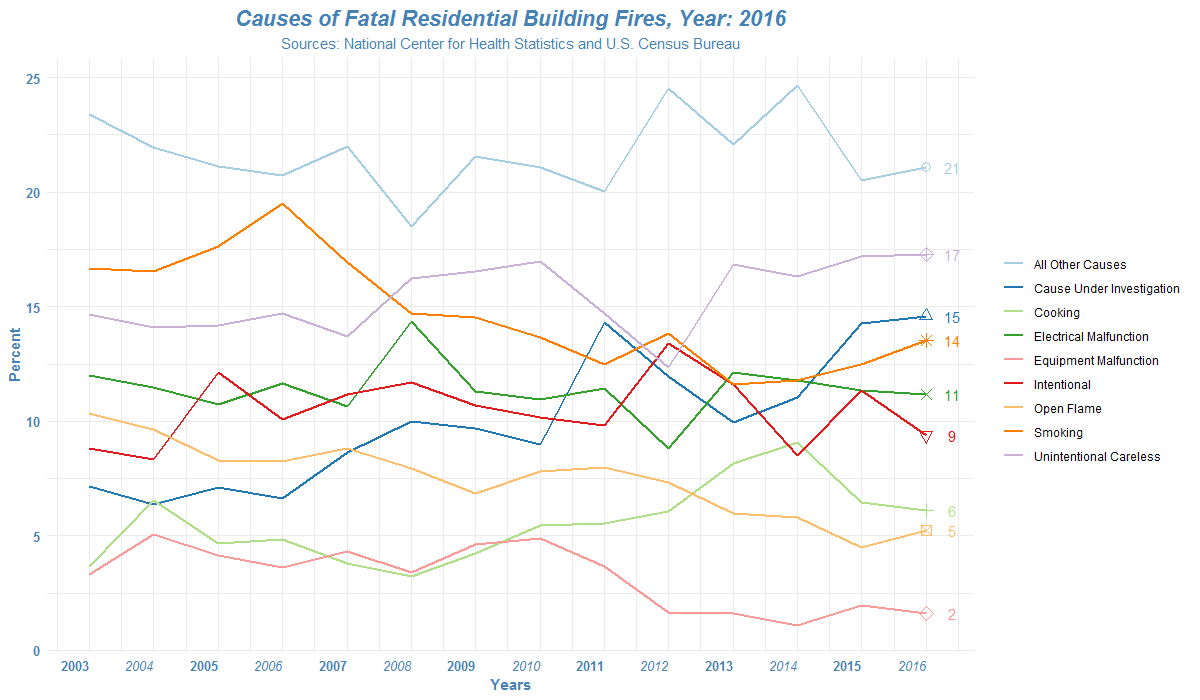
How to read the list of files in R?
When you are working with so many data sets, you don’t want to read each data set, you can read all of data sets at once and access them by a list, that will save you a lot of times.
First of all, let set data directory, it means where you saved your data
# I saved all of data sets in this folder
data_dir <- file.path("U:/Documents/Shiny_Contest_20190315/data")
Let’s check how many data sets in this folder
# list all of file with format csv filenames <- list.files(data_dir, pattern="*.csv", full.names=TRUE) filenames
Now we can read all of files at once, then access the file and checking
# Now we can apply read all of files at once ldf <- lapply(filenames, read.csv) # access and checking the first data set str(ldf[[1]]) 'data.frame': 72 obs. of 5 variables: $ Death_Injuries : Factor w/ 2 levels "Death","Injuries": 1 1 1 1 1 1 1 1 1 1 ... $ Gender : Factor w/ 2 levels "Female","Male": 2 2 2 2 2 2 2 2 2 2 ... $ Age : Factor w/ 18 levels "10_14","15_19",..: 7 10 1 2 3 4 5 6 8 9 ... $ Fire_Death_Percent : num 3.78 2.27 1.7 1.61 2.93 ... $ Fire_Death_Rate_per_Million_Population: num 7.86 4.6 3.42 3.15 5.4 ...
Let’s apply the name for each data instead of access the data by index
# How you asign the name back to the list, so we know which dataset
names(ldf) <- sub('\\..*', '',sub(".*\\/","",filenames))
# Now you can access the file by calling the list plus the name
ldf$`Death and Injuries by Gender and Age 2016`[1:5,]
str(ldf$`Death and Injuries by Gender and Age 2016`)
str(ldf$`Death and Injuries by Gender and Age 2016`)
'data.frame': 72 obs. of 5 variables:
$ Death_Injuries : Factor w/ 2 levels "Death","Injuries": 1 1 1 1 1 1 1 1 1 1 ...
$ Gender : Factor w/ 2 levels "Female","Male": 2 2 2 2 2 2 2 2 2 2 ...
$ Age : Factor w/ 18 levels "10_14","15_19",..: 7 10 1 2 3 4 5 6 8 9 ...
$ Fire_Death_Percent : num 3.78 2.27 1.7 1.61 2.93 ...
$ Fire_Death_Rate_per_Million_Population: num 7.86 4.6 3.42 3.15 5.4 ...
Here is all of code for this post
data_dir <- file.path("U:/Documents/Shiny_Contest_20190315/data")
filenames <- list.files("data", pattern="*.csv", full.names=TRUE)
ldf <- lapply(filenames, read.csv)
names(ldf)<- sub('\\..*', '',sub(".*\\/","",filenames))
# list all of file in data folder
list.files(data_dir)
# list all of file with format csv
filenames <- list.files(data_dir, pattern="*.csv", full.names=TRUE)
# Now we can apply read all of files at once
ldf <- lapply(filenames, read.csv)
# access and checking
str(ldf[[1]])
# How you asign the name back to the list, so we know which dataset
names(ldf) <- sub('\\..*', '',sub(".*\\/","",filenames))
# Now you can access the file by calling the list plus the name
ldf$`Death and Injuries by Gender and Age 2016`[1:5,]
str(ldf$`Death and Injuries by Gender and Age 2016`)
0